Statistical analysis of beer-spoilage microorganisms in 2024
Most frequent beer spoilers | Analysis of findings at the Research Centre Weihenstephan for Brewing and Food Quality, TU Munich (TUM FZW BLQ) about beer-spoiling microorganisms provides a good overview of the status quo, distribution, seasonality and frequency of beer-spoilers.
Though this overview does not present an overall assessment applicable to all breweries, a look at the instances identified at the TUM FZW BLQ provides a fundamental impression of the overall situation of the most frequent problems in brewery microbiology.
Beer-spoilage bacteria
The data from 2024 is presented as the total of instances identified – i.e. batches affected. Several samples of a filling (batch), e.g., are summarised as one individual case. Fermenter or storage tank samples are counted as individual instances, even though these might be combined after the filter cellar into one batch. This inaccuracy has to be inevitably accepted here as production data was not available.
The data represents all analyses of beer-spoilage microorganisms at the TUM FZW BLQ in 2024. The majority of samples originate from Germany, Austria and Switzerland and also come from other EU countries such as France, Italy, the Netherlands and Great Britain, as well as other international data sets.
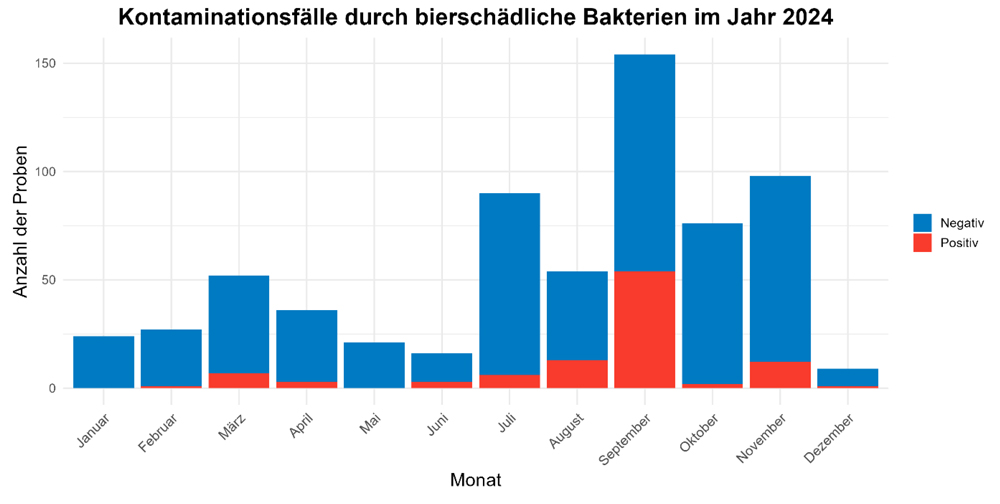
Fig. 1 shows the instances identified in a screening for beer-spoilage bacteria in the months of 2024 subdivided into positive identifications. The screening targeted bacteria of the genera Lactobacillus, Pediococcus, Megasphaera und Pectinatus having a known spoilage potential in beer and mixed-beer drinks. Previous publications provide an overview of the spoilage potential of these microorganisms [1]. As has been the case in previous publications, more instances were noted in the second half of a year, starting with a major upward leap in July. A maximum was noted in particular in September 2024. In addition, proportionally more positive findings were registered during that period [2, 3, 4].
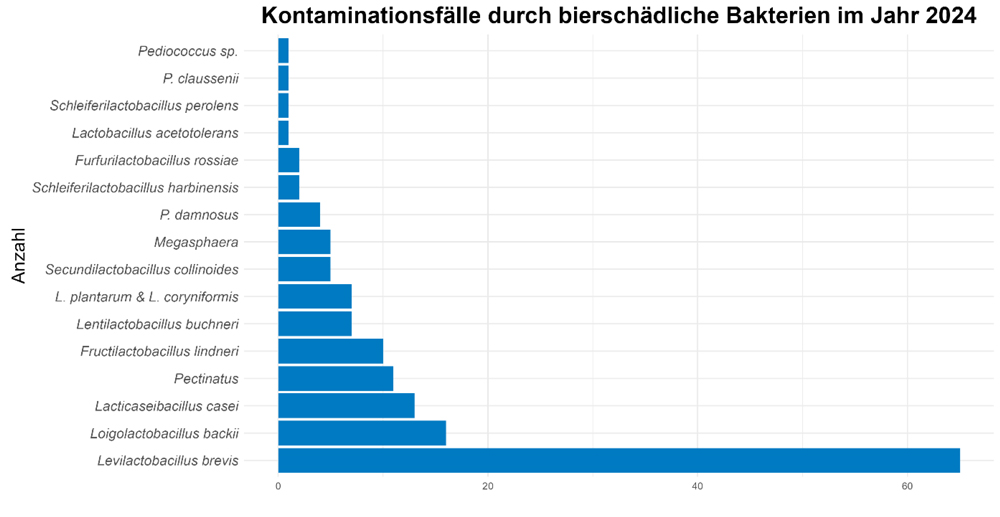
Without any doubt, this can be attributed to the higher output of most breweries during these periods as well as the generally warmer weather and, thus, more rapid biofilm formation in the filling hall. But what types of beer-spoilage bacteria were found in these instances. Fig. 2 shows the distribution of other positive results identified according to species.
As has been reported for many previous years, Levilactobacillus was again the main beer-spoilage bacterium. This obligate beer spoiler is associated with a maximum risk for the product, both as primary as well as secondary contaminant. Loigolactobacillus backii is the next prominent culprit though lagging far behind the former. Other species such as L. buchneri and L. lindneri only account for individual instances. Positive findings of Pectinatus and Megasphaera were identified sporadically in a few breweries in the second half of the year though it should be borne in mind that is difficult to cultivate these species in microbiological routine analysis.
Yeast contaminations
The consequences of a contamination with over-fermenting foreign yeasts such as Dekkera (Brettanomyces) spp. and diastatic Saccharomyces cerevisiae strains (previously referred to as S. cerevisiae var. diastaticus) are obvious: formation of sediments and off-flavours and – even more dangerous – pressure build-up in containers possibly leading to bursting and thus serious injuries. As demand for non-alcoholic products and innovative mixed beverages is on the increase, breweries are facing ever new technological challenges; an increasing spectrum of (potentially) product-spoiling microorganisms is developing. Even common culture yeast in beer ready for sale poses a major risk in non-alcoholic beer and the search for fermentable yeasts is becoming ever more important within this classification [5].
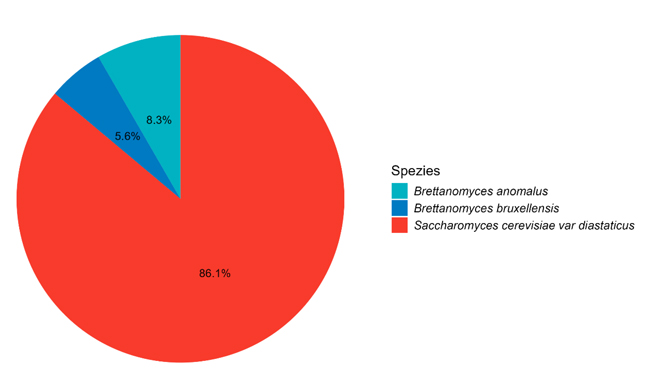
It is difficult to unambiguously identify the latter in this context due to sample coding. Therefore, just an overview of yeasts which can spoil practically all products is given here. Fig. 3 shows instances of over-fermenting yeasts and the breakdown of species. A total of 123 samples with over-fermenting yeast were recorded.
Suspicious instances of beer-spoiling bacteria, as well of over-fermenting yeasts, are on the increase, so is the proportion of positive findings at the beginning of the second half of the year. Only individual instances of Dekkera spp. were identified in the course of the year; diastatic Saccharomyces cerevisiae account for by far the largest share of incidents recorded by us.
MALDI-ToF analysis
Since our last corresponding reporting in 2013 [4], use of Matrix Assisted Laser Desorption Ionisation Time of Flight-Mass Spectrometry (MALDI-ToF) for identification of microorganisms has been introduced. This method, compared to real-time PCR systems, has no result-oriented expectations (“open system”). A real-time system reacts positively only to the species that is actively sought by the specialised, specific primers. MALDI-ToF can successfully identify a microorganism as long as its spectrum is stored in the (ever growing) database. Both systems naturally have downsides and are not suited for certain sample matrices or incidents. However, the MALDI-ToF data permits the establishment of an overall picture of microorganisms detected as the search is not specifically targeted to beer spoilers.
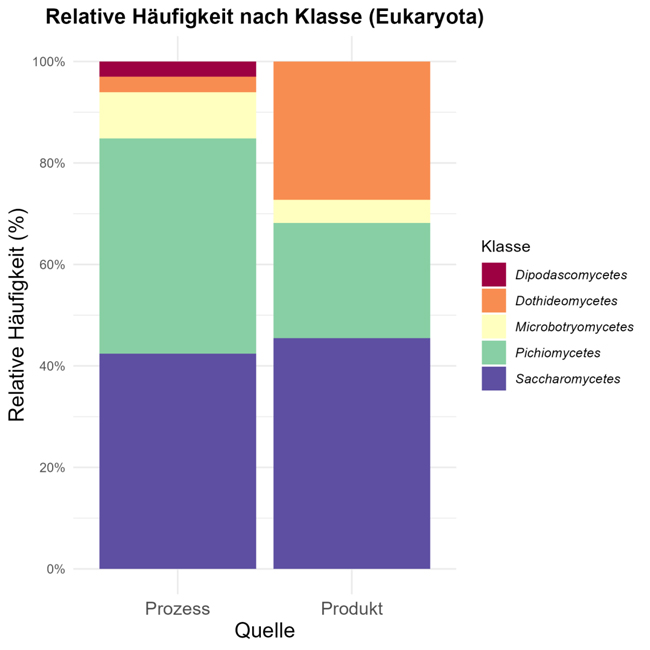
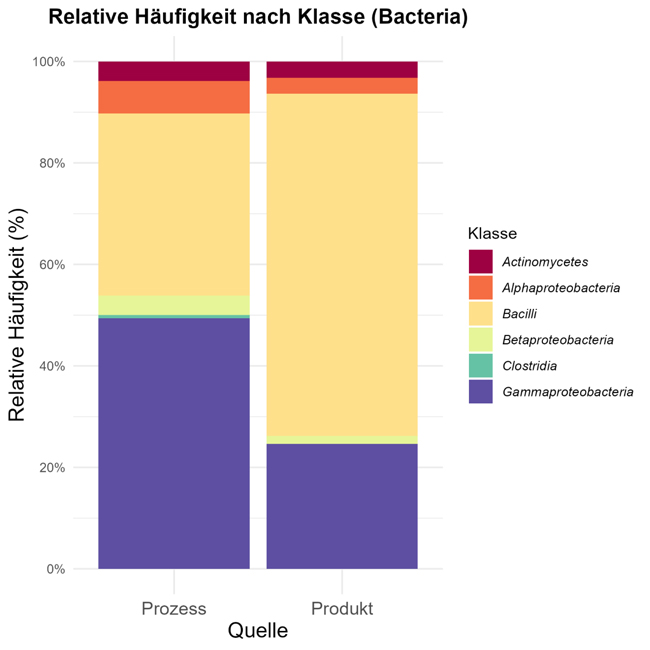
Figs. 4 and 5 show overviews of microorganisms found in samples (fermenters, yeast deposits, storage tanks, pressure tanks, filling hall, filled containers) at the taxonomic level of families and thus map the microorganism associated with the brewing process, together with classical culture and harmful yeasts and beer-spoilage microorganisms.
In view of the fact that the method does not focus on specific results and its simple and rapid execution, many investigations of intermediate products and cultivations on various nutrient media are included. The nutrient medium used naturally also has an influence of the spectrum of microorganisms growing. The image is subdivided into process samples and filled containers (bottles, cans, barrels) and is representative of a total of 1056 instances included in the statistic, which could be associated with process steps and products.
The beer-spoilage species of Lactobaccillae frequently found are classified under Bacilli, together with other Bacillus species that are detected in many samples in view of their very resistant spores. However, these species do not pose a known spoilage risk in finished beverages.
Gamma proteobacteria, such as Pseudomonas or enterobacteria, are identified in particular in intermediate products, e.g. process waters or samples from surroundings. They are able in find niches there as they require few nutrients and have a high potential for biofilm formation. Among Alphaproteobacteria, known representatives such as Acetobacter and Gluconobacter are found in the beverage area.
As was to be expected, cultured yeasts (class of Saccharomycetes - Wickerhamomyces, Hanseniaspora and Torulapsora) are yeasts frequently identified as large numbers of these are present in breweries. Other species such as Candida, Wickerhamomyces or Rhodotorula (Microbotryomycetes) are also frequently detected in rinse waters or samples from the surroundings of equipment.
Beverage-relevant yeasts, such as Pichia, Dekkera and Candida, are representatives of the class of Pichiomycetes. In the finished product, some bacterium species are found in biofilms at the filler. They are not associated with any spoilage potential, they die in the beverage or their spores (e.g. Bacillus sp.) survive for a long time, without germinating.
Conclusion
L. brevis remains the top beer spoiler as primary and secondary contaminants among beer-spoilage bacteria. Other species are much rarer or can be traced back to few instances in individual operations. Microbiologically suspicious instances and positive findings are mainly found at the beginning of the second half of a year (August, September and October), the same applies to detection of over-fermenting yeasts. Evaluation of microorganisms identified using MALDI-ToF identify both beer spoilers as well as microorganisms in the surroundings of equipment. These do not necessarily spoil the product but are associated with the process of beer brewing.
References
- Hutzler, M.; Müller-Auffermann, K.; Koob, J.; Riedl, R.; Jacob, F.: “Beer spoiling microorganisms – a current overview”, BRAUWELT International, 1, 2013, pp. 23–25.
- Back, W.; Breu, S.; Weigand, C.: “Infektionsursachen im Jahre 1987”, BRAUWELT, 31/32, 1988, pp. 1358–1362.
- Hutzler, M.; Koob, J.; Grammer, M.; Riedl, R.; Jacob, F.: „Statistische Auswertung der PCR-Analysen bierschädlicher Bakterien in den Jahren 2010 und 2011“, BRAUWELT, 18–19, 2012, pp. 546–547.
- Koob, J.; Jacob, F.; Grammer, M.; Kleucker, A.; Riedl, R.; Hutzler, M.: “PCR-Analysen bierschädlicher Bakterien 2012 und 2013”, BRAUWELT, 10, 2014, pp. 288–290.
- Hutzler, M.; Riedl, R.; Koob, J.; Jacob, F.: “Fermentation and spoilage yeasts and their relevance for the beverage Industry – A Review”, BrewingScience, 65, 2012, pp. 33–52, https://doi.org/10.23763/BrSc12-07hutzler.